Table of Contents:
- Strengths of E. coli Expression Systems
- Common Challenges in E. coli Expression
- Solutions and Best Practices
- Advanced Applications of E. coli in Research
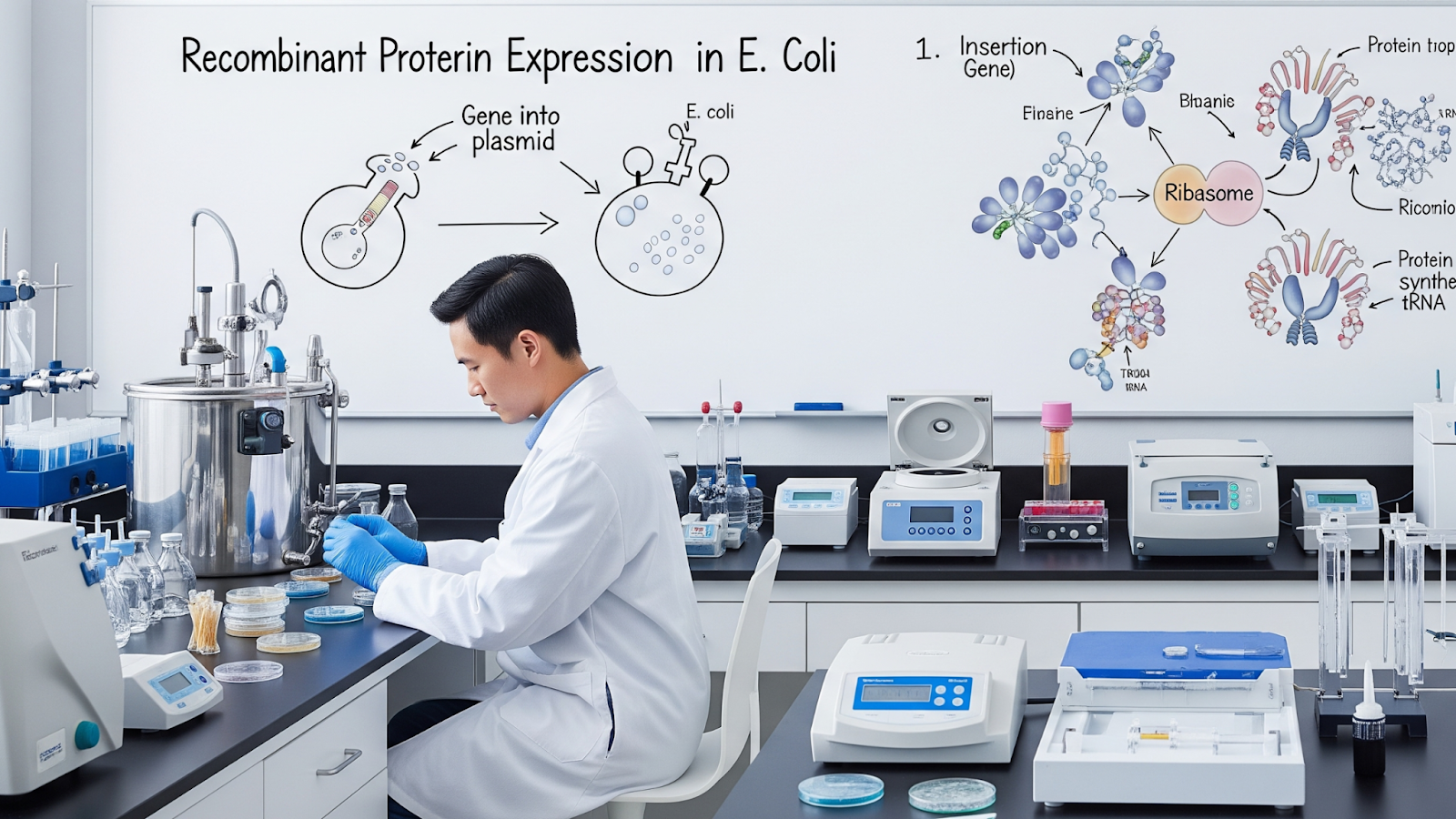
Recombinant protein expression enables researchers and scientists to produce recombinant proteins in a controlled environment. Escherichia coli remains the most widely used host due to its fast protein production, cost efficiency, and the ease with which its plasmids and genetic elements can be manipulated to optimize expression.
E. coli, as a host system, provides researchers with a reliable and scalable approach for large-scale recombinant protein production, suitable for both fundamental studies and translational applications.
For example, the CUL1 recombinant protein is a core component of SCF-type E3 ubiquitin ligase complexes. These complexes regulate protein degradation pathways that play a critical role in controlling the cell cycle.
CUL1 is not an abundant protein in cells. Recombinant expression makes it accessible in the quantities needed for research.
Strengths of E. coli Expression Systems
Cost-Effectiveness
E. coli has a low cultivation cost, and its growth media are inexpensive. E. coli cells multiply rapidly under optimal conditions, with a doubling time of about 20 minutes. The increased number of cells increases the production of proteins.
Scalability
E. coli seamlessly scales from small-scale experiments to industrial production. Laboratory protocols for protein expression are easily transferable to larger fermentation systems with minimal changes.
Genetic Manipulation
Researchers can use plasmid vectors, promoters, ribosome binding sites, and codon optimization strategies to insert, modify, or optimize genes within E. coli easily.
Established Protocols
From expression to purification, there are standardized, well-documented protocols for every stage of protein production. Induction systems, affinity tags, and purification methods are widely available and validated across laboratories worldwide.
Common Challenges in E. coli Expression
Inclusion Bodies
When large amounts of recombinant protein are produced and chaperone support is not enough, many newly formed protein chains do not fold into the correct 3D shape.
The hydrophobic regions of unfolded proteins are exposed and stick to each other. As a result, these unfolded proteins clump together, forming inclusion bodies. "This is one reason why recombinant human protein expression in E. coli may require additional refolding steps.
Endotoxin Contamination
E. coli naturally produces lipopolysaccharides (LPS) called endotoxins. These contaminants get purified with recombinant proteins and must be removed.
Lack of Post-Translational Modifications
E. coli lacks enzymatic systems for glycosylation and other post-translational modifications, making it challenging to produce fully functional versions of some recombinant human proteins.
Solutions and Best Practices
Solubility Tags
Inclusion body formation can be prevented with solubility-enhancing fusion tags such as GST, MBP, or SUMO. These tags maintain the solubility of the target protein and prevent its aggregation. They can later be cleaved off to obtain the native form of the protein.
Codon Optimization and Engineered Strains
Codons guide the cell in assembling amino acids into proteins in the correct order. E. coli contains some rare codons that are not used frequently. It may struggle to translate them, which can lead to slow or stalled protein synthesis, misfolded proteins, or truncated proteins.
Codon optimization is done to replace these rare codons with the codons preferred by E. coli.
Normal strains sometimes fail to produce or fold complex proteins correctly. Engineered strains, such as Rosetta, SHuffle, and Origami, are designed to address these specific challenges.
Refolding and Chaperone-Assisted Folding
Sometimes, it is not possible to avoid inclusion bodies. In such cases, proteins can be solubilized under denaturing conditions. They are then carefully refolded to ensure functionality with the help of co-expression of molecular chaperones.
Endotoxin Removal
Endotoxins (LPS) are harmful in therapeutic and diagnostic applications. Methods such as affinity resins, detergent washes, or chromatography help capture LPS during purification.
When to Choose E. coli Over Yeast or Mammalian Systems
E. coli may not be the best choice for proteins requiring extensive post-translational modifications, complex folding, or secretion. This is where yeast (Pichia pastoris) or mammalian expression systems can be beneficial.
Advanced Applications of E. coli in Research
Structural Biology
Researchers use high-yield recombinant proteins from E. coli in X-ray crystallography, cryo-EM, and NMR studies to reveal protein mechanisms, binding sites, and conformational changes.
Drug Discovery
Recombinant proteins from E. coli serve as accessible targets for high-throughput screening, ligand-binding assays, and inhibitor testing, accelerating therapeutic development.
Synthetic Biology
E. coli functions as a chassis for designing metabolic pathways, biosensors, and genetic circuits to support the production of biologics, novel biomaterials, and biofuels.
Protein Engineering
The genetic tractability of E. coli makes it a preferred system for site-directed mutagenesis, directed evolution, and optimization of protein stability, solubility, and activity.
Industrial Biotechnology
Engineered E. coli strains are used in large-scale production of insulin, antibody fragments, enzymes, and other recombinant bioproducts with clinical and commercial value.
Conclusion
E. coli is a cost-effective host for recombinant protein expression. Researchers can use engineered strains, solubility tags, and purification strategies to address challenges such as inclusion bodies, misfolding, or endotoxins.












